OpenFold2
Replicating AlphaFold2 in the dark
Introduction
AlphaFold2 is a monumental step towards making biology an engineering discipline. Ability to predict protein structures with high accuracy and reliably finally gives humanity the keys to the living matter. We believe, that this algorithm is too important to be unavailable for modifications and improvements.
In this work we are replicating complete AlphaFold2 algorithm. However at the moment of writing this text only surface-level details are known about this algorithm, which make replication effort non-trivial. To overcome this challenge we use the dataset-driven tactics to build the complete model.
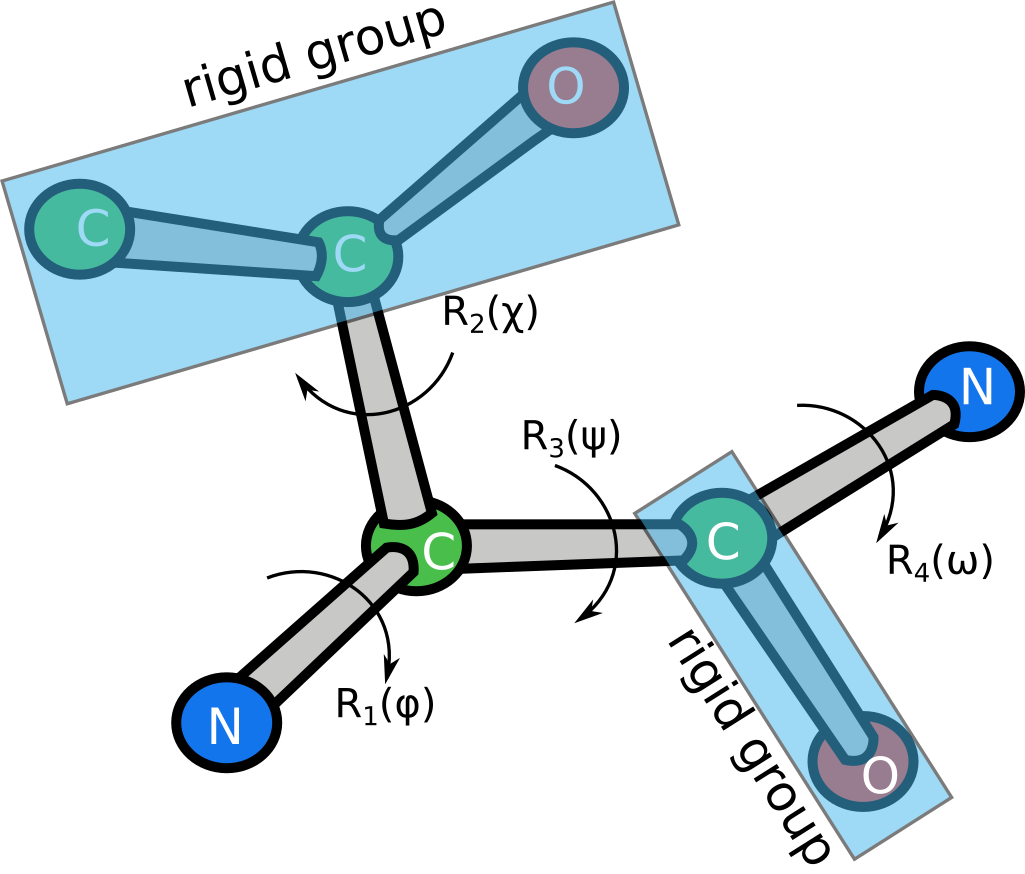
The key idea is to break the whole AlphaFold2 algorithm in several parts: SE(3) equivariant part, MSA part, structural module, possible unsupervised pretraining. We then design a dataset that examplifies the rules each part tries to capture during the training and build a model for this dataset.
This approach makes replicating complex composite models like AlphaFold2 more tractable. We also can explore failure modes of each part of the model without training it on the complete dataset of protein MSAs and structures.
Iterative SE(3) transformer
Predicting particle coordinates using iterative se(3) transformer.