MSA
Code
Introduction
Multiple sequence alignment is a key input of AlphaFold2 algorithm. However, the size of the attention matrix of the transformer that operates on MSA is $~N^2 M^2$, where $N$ is the sequence length and $M$ is the number of sequences in the alignment. This is prohibitively costly even for the DeepMind group.
In this chapter, we show how to efficiently incorporate MSA into structure prediction. The key observation is that alignment of multiple sequences assumes that they have similar structures. Also, the correlation between the mutations in the MSA stems from the proximity of these residues in the protein structure.
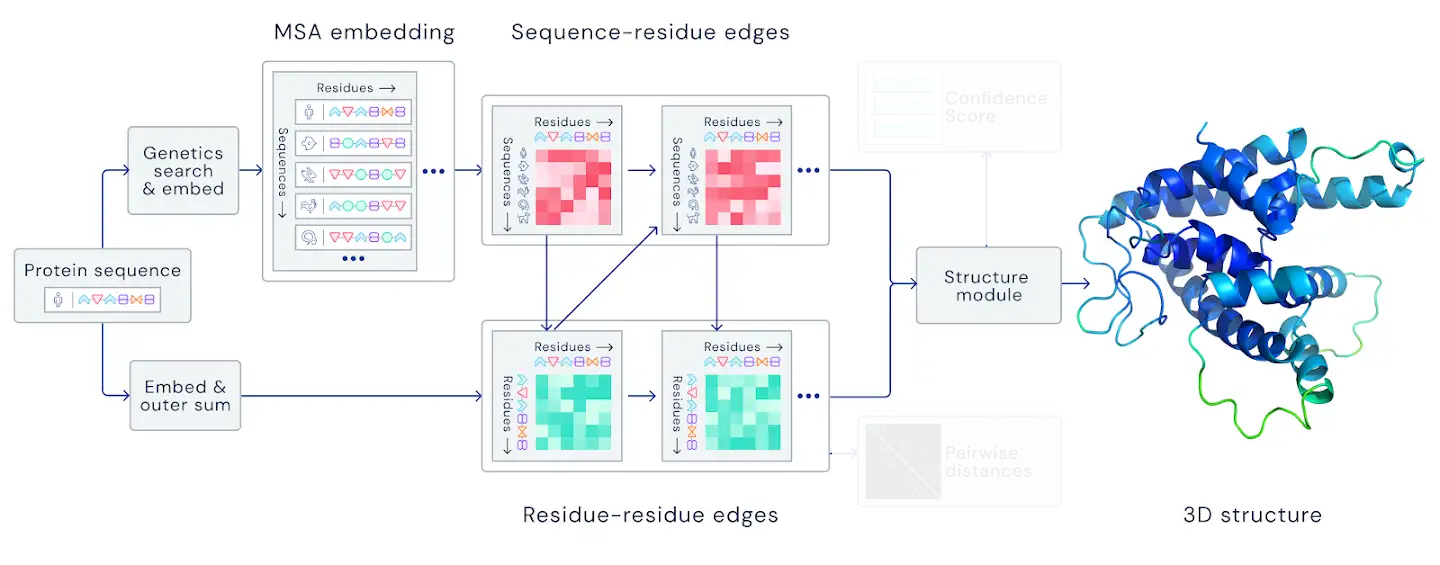
Figure 1: MSA part of AlphaFold2.
Dataset
This dataset serves mainly one purpose: to verify that the algorithm can learn structures of proteins from MSA alone. The sequence, that is served to the structural part is random, while all the information about the arrangement of patters is contained in the MSA. Otherwise this dataset closely resembles the one we used in the previous chapter.
Similar to the previous dataset, we generate an MSA (Figure 2A), consisting of fragments(Ala) and patterns(Gly). Afterwards, we generate displacements of patters and insert different amino-acids (R/N) at the beginning of a patterns, that correspond to the displacements. However in this case each of them are inserted in specific sequence, while the first one remains random. This way we ensure that all the information about the structure can only be extracted while observing MSA and not the first sequence.
Then next two setps are the same as in the previous chapter (Figure 2B, Figure 2C). Finally, Figure 2D shows samples from the toy MSA dataset. Additionally we increased the size of the dataset and the length of the sequences to ensure, that the model can not memorize the first random sequences.
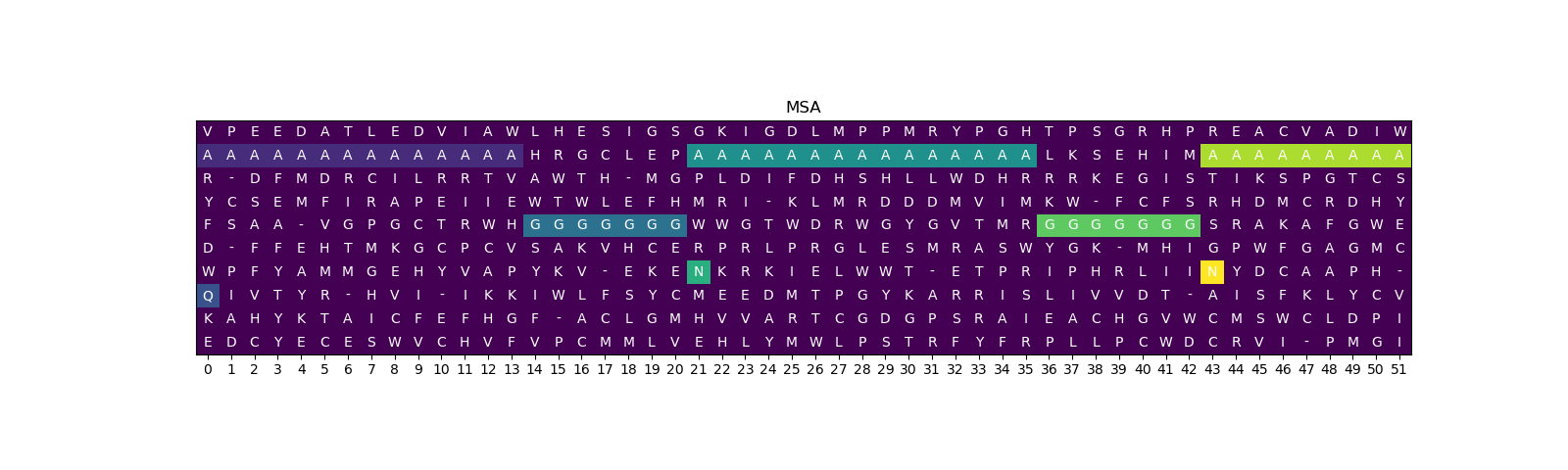
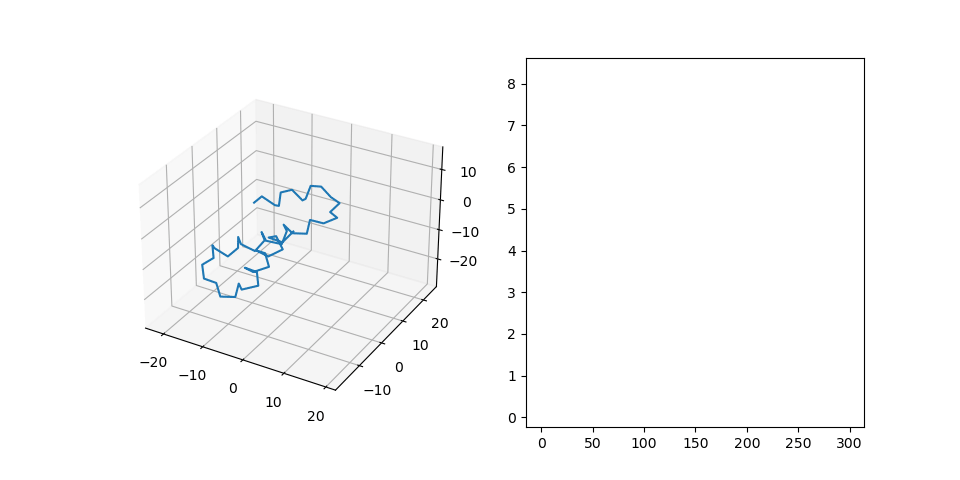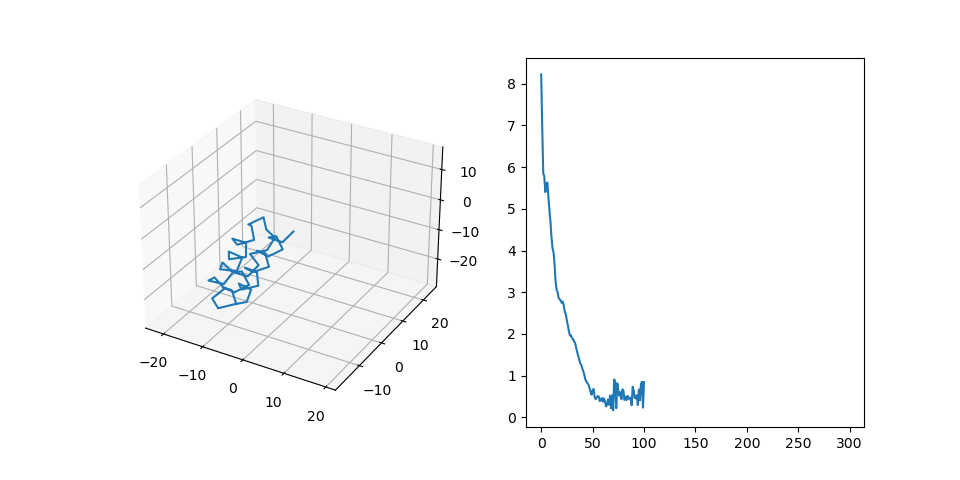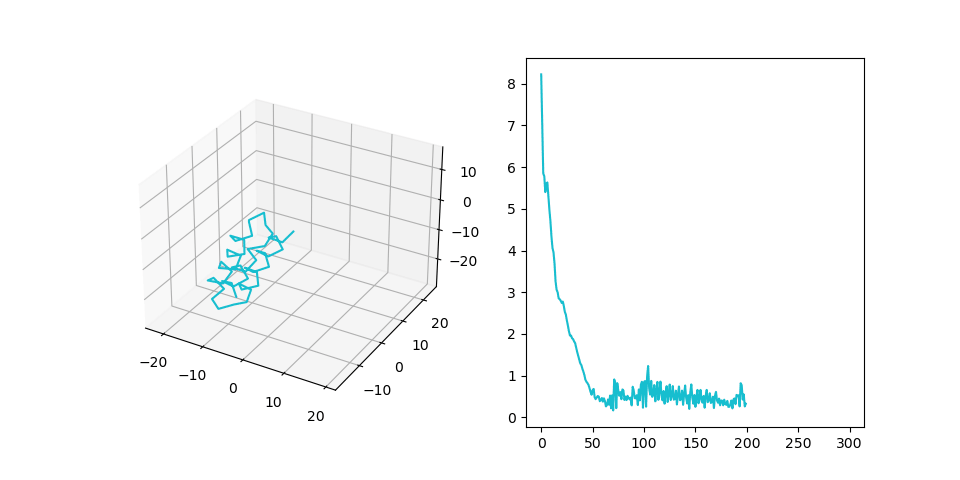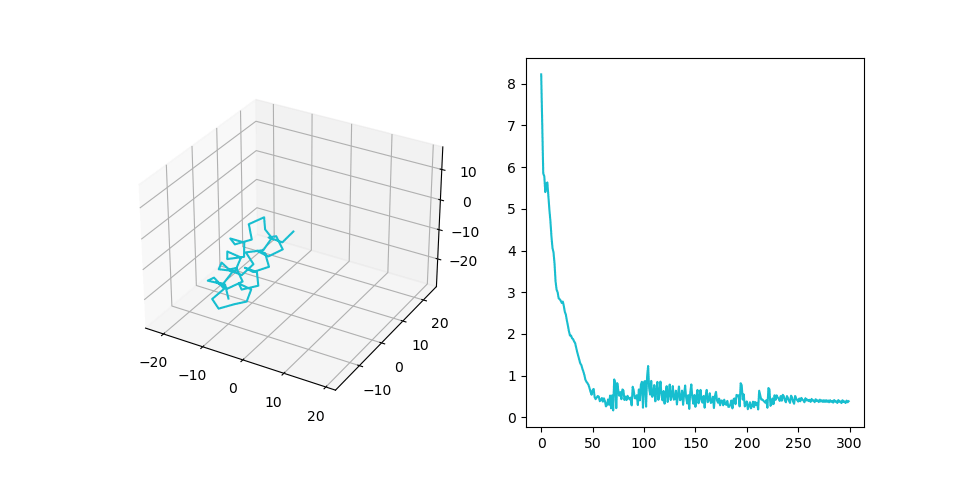
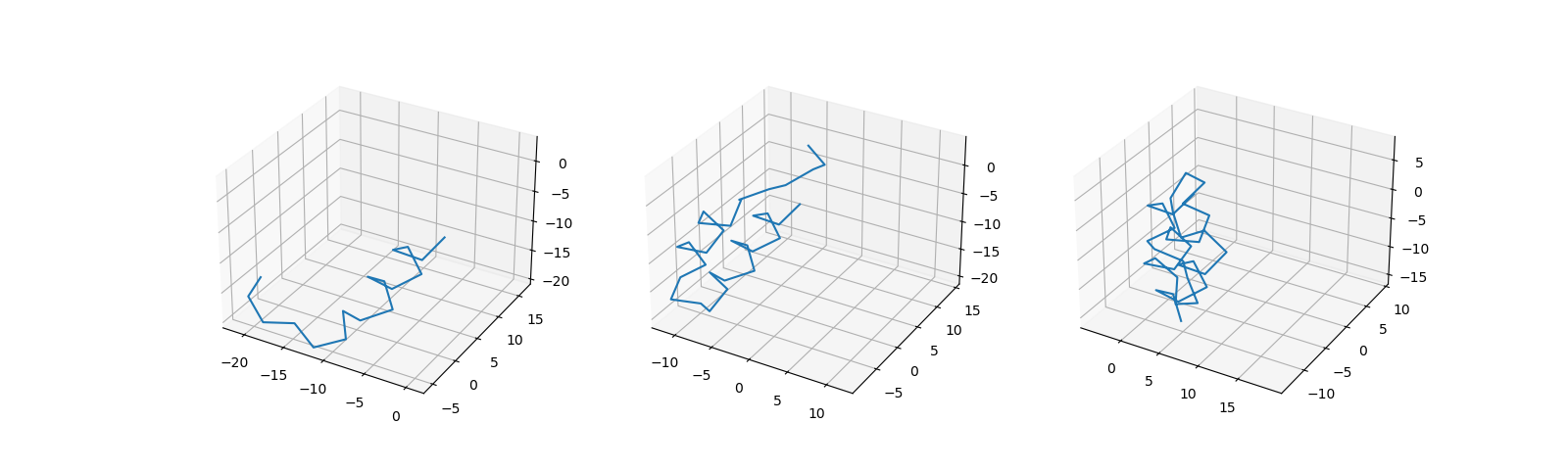
Figure 2: MSA dataset generation steps.
Model architecture
Figure 3 gives shematic representation of the model architecture. Similar general idea was first published in the paper "MSA Transformer"[1], where Roshan Rao et al. used unified attention over sequences, coupled with the column attention to circumvent the whole attention matrix computational cost. However in AlphaFold2 the attention over sequences correspond to the structure of the protein, therefore we use distance matrix between centers of rigid bodies to compute row attention matrix: $$ att_{ij} = \frac{1}{12.0}ReLU\left( 12.0 - |\mathbf{x}_i - \mathbf{x}_j|\right) $$ where $\mathbf{x}_i$ are the coordinates of $i$th node after the SE(3) transformer block.
Additionally we need two-way communication between structural and MSA parts of the model. We use the output features of the MSA transformer block corresponding to the first sequence in the MSA as the node input to the next SE(3) transformer block.
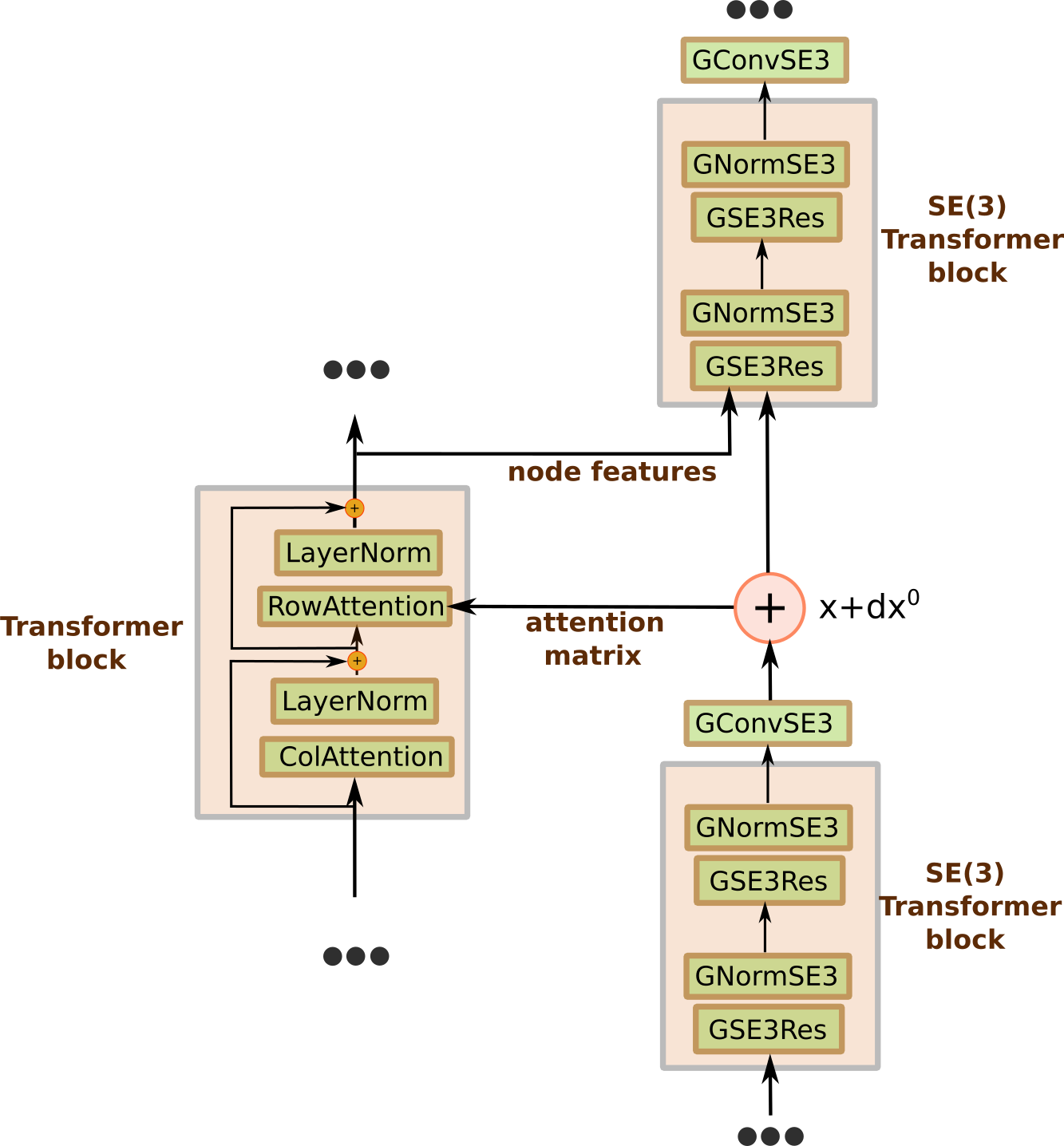
Figure 3: MSA transformer model architecture.
Results
In this experiment we paid additional attention to the overfitting problem. Our goal here is to check whether the model can propagate information from the MSA part of the input to the SE(3) part. Therefore we have to make sure, that the algorithm does not simply memorize random amino-acid sequences that we generated. We incresed the size of the dataset (1000 examples, sequence length 40-80, num sequences in MSA 10) and decreased the size of the model. After training for 100 epochs we have the following result:| # | Train | Test |
|---|---|---|
| Epoch 0 | 6.90 | 8.36 |
| Epoch 100 | 1.98 | 2.98 |
Citations
- 1. Rao, Roshan, et al. "Msa transformer." bioRxiv (2021).